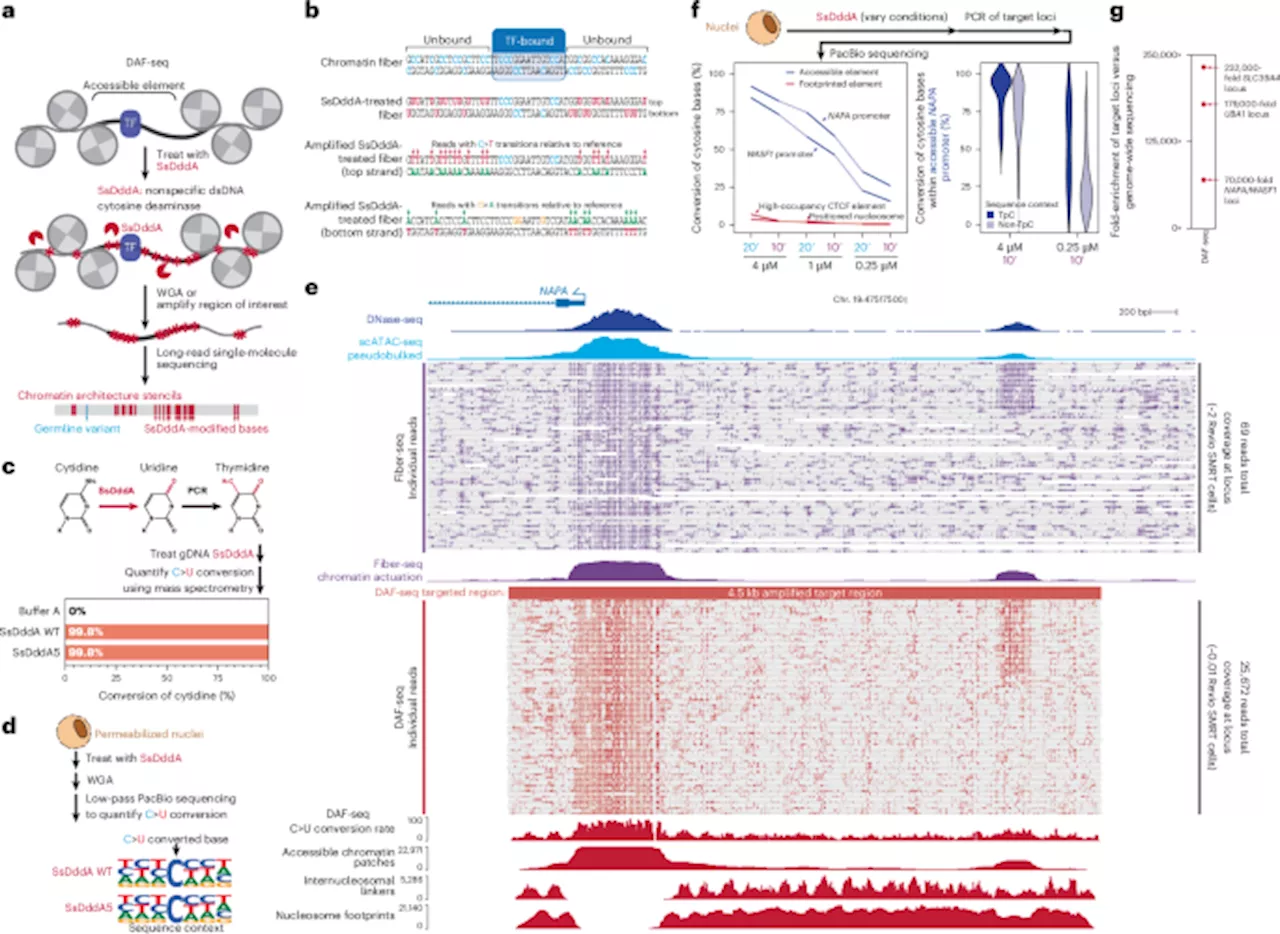
Researchers at the University of Washington have developed a groundbreaking sequencing method called Deaminase-Assisted single-molecule chromatin Fiber sequencing, or DAF-seq. This innovative technique offers unprecedented insights into gene regulation by mapping single-cell chromatin fiber architectures. The findings promise to advance understanding of how proteins interact with genetic material within individual cells, particularly in diploid organisms, where such interactions have been less well understood.
DAF-seq enables researchers to achieve near-nucleotide resolution in single-molecule footprinting. This allows for the simultaneous profiling of chromatin states and DNA sequences, shedding light on how proteins co-bind along chromosome-length chromatin fibers. The technique has revealed significant heterogeneity in protein occupancy among different haplotypes and cells.
The study demonstrated that single-cell DAF-seq generates comprehensive chromosome-length protein co-occupancy maps, covering 99% of each individual cell’s mappable genome. The results indicate extensive chromatin plasticity, with chromatin actuation varying by 61% between haplotypes within a cell and 63% across different cells. This variability suggests a complex regulatory landscape where cooperative protein interactions occur in a distance-dependent manner, akin to the behavior of cohesin-mediated loops.
A.B. Stergachis, a lead researcher involved in the project, emphasized the significance of these findings. “DAF-seq not only provides a detailed view of chromatin architecture but also highlights the functional impact of somatic variants and rare chromatin epialleles,” he stated. The insights gained could have implications for understanding genetic variation in health and disease.
The research also acknowledges the collaborative efforts of various institutions and funding bodies, including the Burroughs Wellcome Fund and the Chan Zuckerberg Initiative. The study received support from the National Institutes of Health under several grant awards, which enabled the development of this innovative method.
This advancement in chromatin research opens new avenues for exploring gene regulation and its implications for human health. By characterizing protein occupancy with single-nucleotide precision, DAF-seq stands to enhance the understanding of complex genetic interactions and their consequences in various biological processes.
The full findings are detailed in the research published by the team, which can be accessed through their GitHub repository. As ongoing research continues to unravel the complexities of genome regulation, techniques like DAF-seq are poised to play a crucial role in the future of genomic studies.