BREAKING: A revolutionary deep learning model developed by a team from KU Leuven has just transformed the way scientists analyze fruit tissue microstructure. This breakthrough was published in Plant Phenomics on July 5, 2025, unveiling a fully automated framework that dramatically enhances the accuracy of 3D imaging for apples and pears.
The innovative model outperformed traditional 2D approaches, achieving an impressive Aggregated Jaccard Index (AJI) of 0.889 for apples and 0.773 for pears. This advancement is crucial for researchers who depend on precise tissue segmentation for understanding vital metabolic processes in plants.
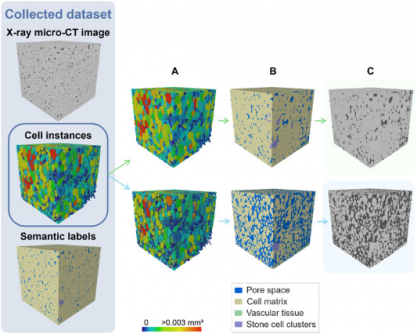
Traditional microscopy techniques have long been constrained by extensive sample preparation and limited viewing areas. X-ray micro-CT has introduced non-destructive 3D imaging capabilities. However, challenges such as low image contrast and overlapping features have made it difficult to quantify tissue morphology accurately. The newly developed deep learning model addresses these limitations, offering a robust solution for the complex task of segmenting various plant tissue types.
Utilizing a 3D panoptic segmentation framework, the model effectively performs both instance and semantic segmentation. It accurately predicts the individual components of fruit tissue, including parenchyma cells, vascular tissues, and stone cell clusters. The model was trained on datasets of apples and pears, employing advanced synthetic data augmentation techniques. This groundbreaking approach enables a complete labeling of fruit tissue microstructure in real-time, marking a significant leap forward in plant research.
Visual validation confirmed that the model accurately detects vascular bundles in popular apple varieties such as ‘Kizuri’ and ‘Braeburn,’ showcasing its potential for practical applications in agriculture. Its ability to segment stone cell clusters with a Dice Similarity Coefficient (DSC) of up to 0.90 highlights the model’s precision and reliability.
This deep learning framework not only accelerates data analysis but also reduces the manual labor traditionally associated with tissue characterization. Researchers will now be able to explore how microscopic structures influence water, gas, and nutrient transport, leading to insights that can enhance fruit texture, storability, and resistance to disorders like browning.
Looking ahead, the model presents a scalable solution for studying various crops, enabling scientists to investigate tissue development, ripening, and stress responses across different agricultural contexts. Its compatibility with existing X-ray micro-CT equipment ensures that this cutting-edge technology can be readily adopted by plant scientists worldwide.
The research was funded by the Research Foundation – Flanders (FWO) and KU Leuven, and it signifies a major step forward in integrating artificial intelligence into plant anatomy and food science research.
As the agricultural community grapples with challenges posed by climate change and food security, this new technology offers hope. It provides a powerful tool for researchers to better understand the intricate relationships within plant tissues, ultimately leading to improved agricultural practices and crop resilience.
Stay tuned for further developments as this technology continues to evolve and impact the future of plant science.